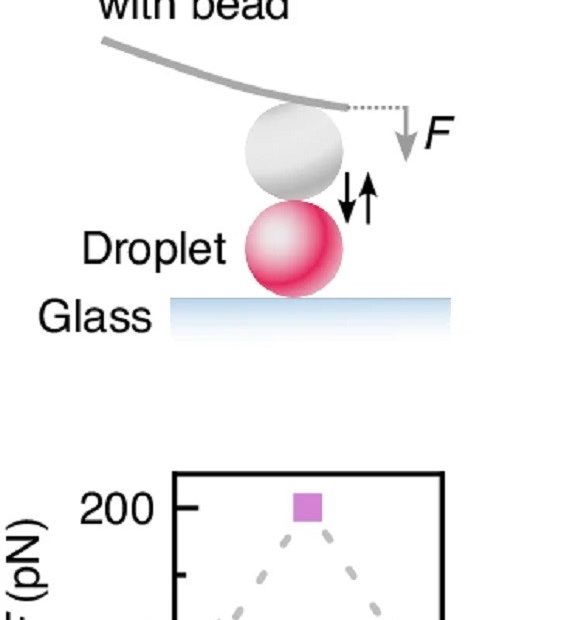
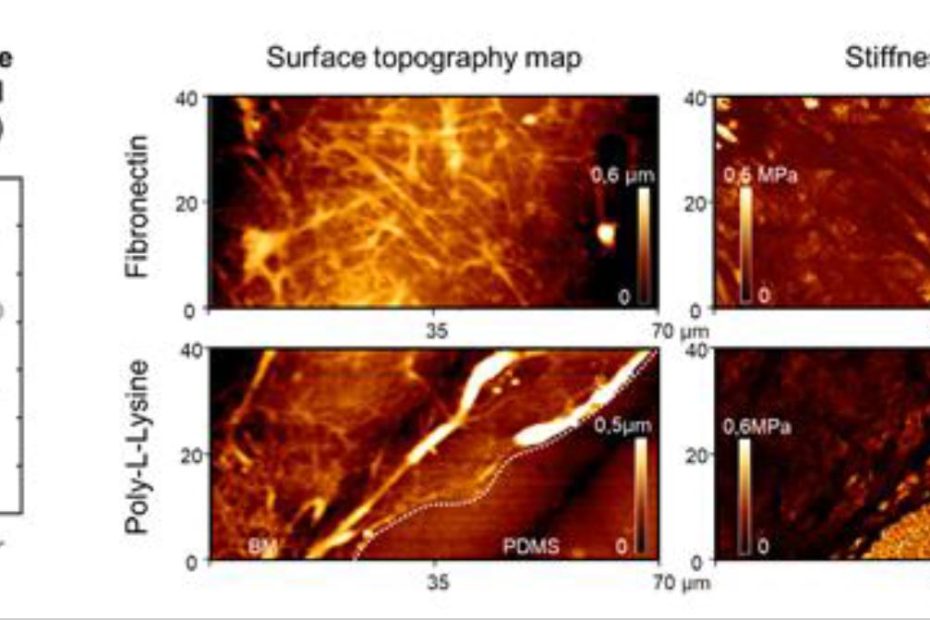
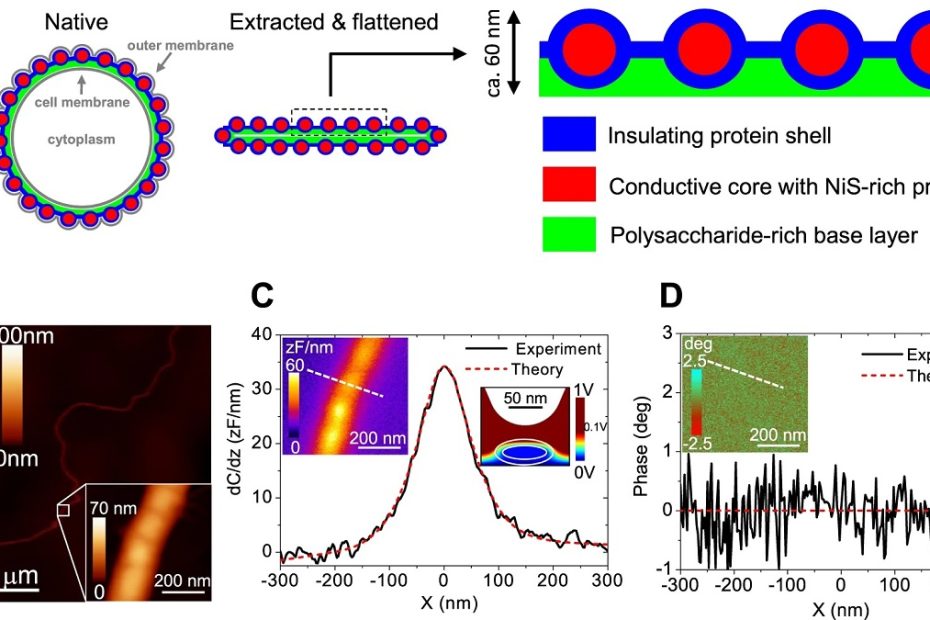
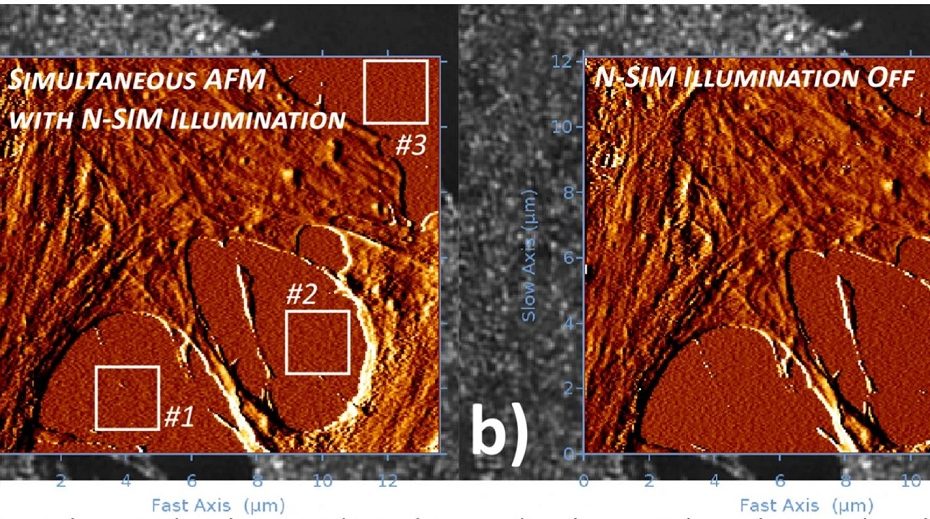
Deformable microlaser force sensing
Mechanical forces are key regulators of cellular behavior and function, affecting many fundamental biological processes such as cell migration, embryogenesis, immunological responses, and pathological states.… Read More »Deformable microlaser force sensing