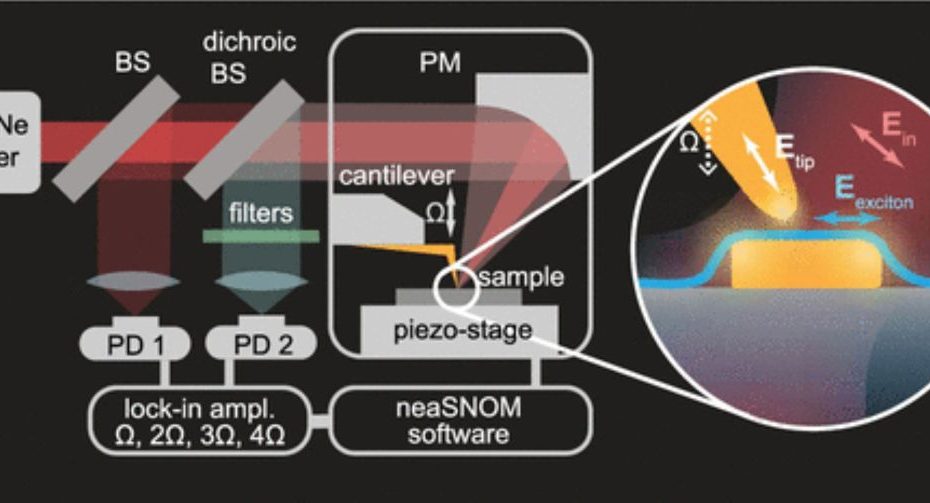
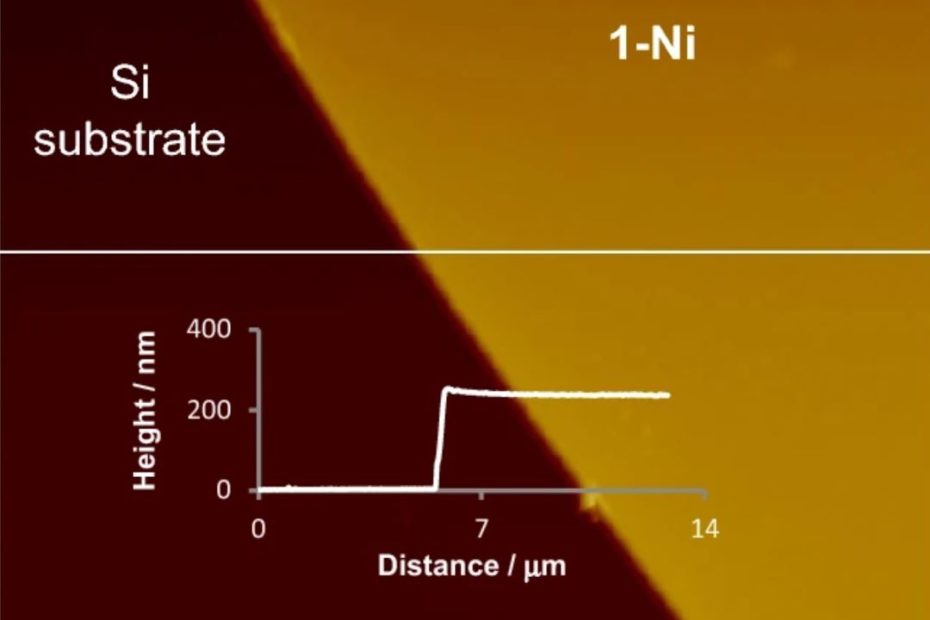
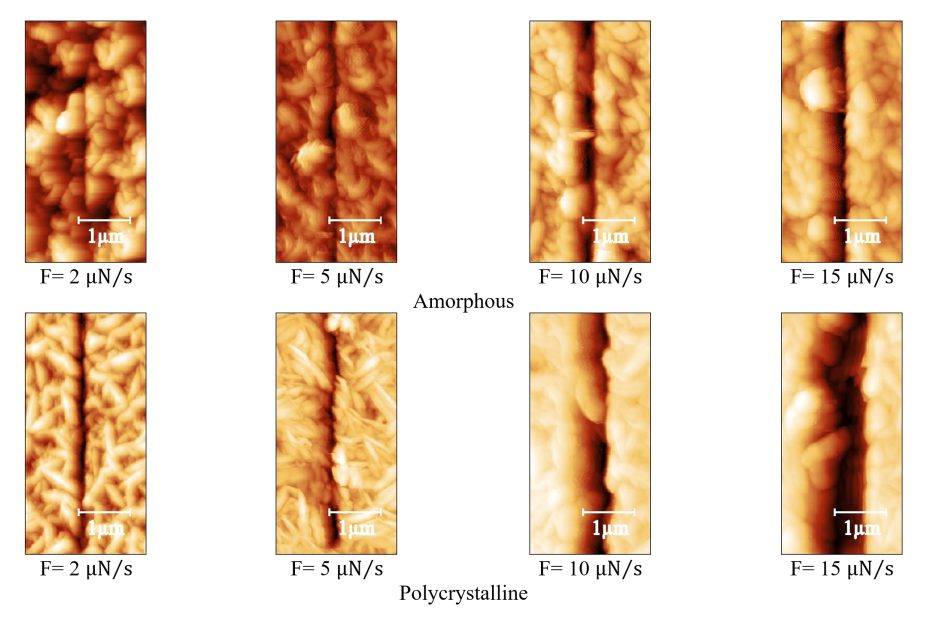
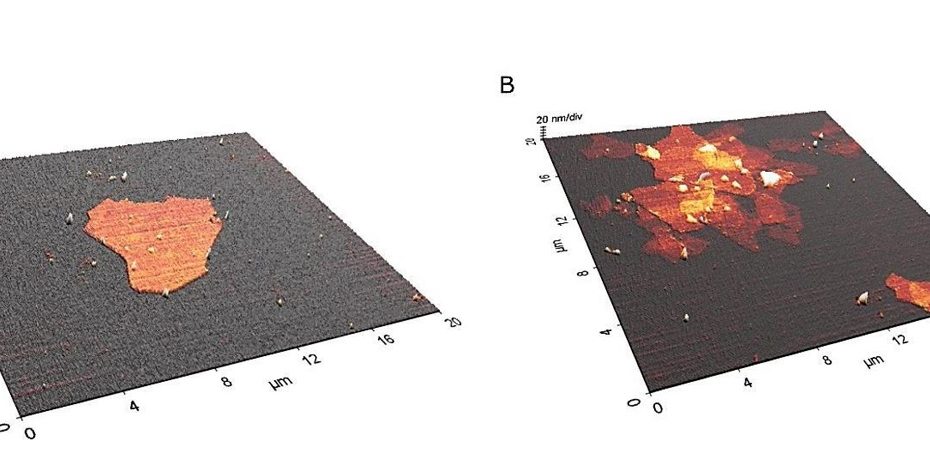
Enhanced Exiton-Plasmon Interaction Enabling Observation of Near-Field Photoluminescence in a WSe2-Gold Nanoparticle Hybrid System
Monolayer transition metal dichalcogenides, such as tungsten diselenide, have recently attracted considerable attention due to their reduced dielectric screening and direct bandgap, which result in… Read More »Enhanced Exiton-Plasmon Interaction Enabling Observation of Near-Field Photoluminescence in a WSe2-Gold Nanoparticle Hybrid System