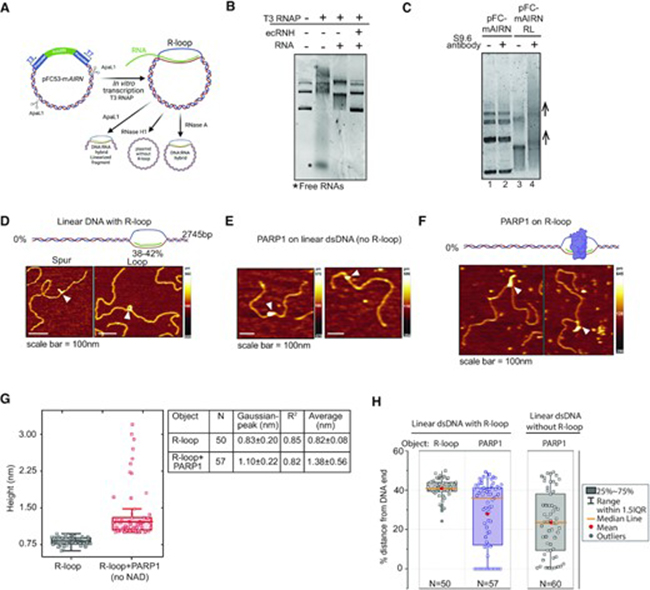
In a landmark study published in Nucleic Acids Research, a team of scientists has uncovered a novel role for PARP1, a key DNA repair enzyme, in regulating R-loops—three-stranded nucleic acid structures formed during transcription. While R-loops play essential regulatory roles, their accumulation can lead to DNA damage and genome instability. The study shows that PARP1 directly binds to R-loops and promotes their resolution, adding a new layer to our understanding of how cells safeguard genome integrity.
This interdisciplinary work was carried out by a diverse team of researchers led by:
Natalie Laspata, Parminder Kaur, Sofiane Yacine Mersaoui, Daniela Muoio, Zhiyan Silvia Liu, Maxwell Henry Bannister, Hai Dang Nguyen, Caroline Curry, John M. Pascal, Guy G. Poirier, Hong Wang, Jean‑Yves Masson, and Elise Fouquerel.
Their affiliations span institutions across the USA and Canada, including the University of Pittsburgh, Thomas Jefferson University, Laval University, University of Minnesota, and Université de Montréal. This collaboration brings together expertise in RNA biology, DNA repair, biochemical reconstitution, and structural imaging.
🔬 Read the full article here:
Nucleic Acids Research, Volume 51, Issue 5, 21 March 2023, Pages 2215–2237, https://doi.org/10.1093/nar/gkad066
Published: 16 February 2023
AFM enabled the researchers to directly observe R-loop structures and detect how PARP1 binding changed their configuration, offering clear visual evidence of PARP1’s resolving activity. That’s where the PPP-FMR AFM probe from Nanosensors came into play.
The PPP-FMR probe is optimized for force modulation mode, which is ideal for mapping soft biological materials. With a sharp tip radius (<10 nm) and a finely tuned spring constant (~2.8 N/m), it provides the sensitivity needed to distinguish between DNA strands, RNA hybrids, and protein complexes.In this study, PPP-FMR probes were essential for imaging plasmid DNA containing R-loops, and for observing how PARP1 binding altered their shape or frequency. Their compatibility with fluid imaging conditions ensured the biomolecules remained in their native state throughout the experiment.Thanks to the precision and reliability of the PPP-FMR probes, the research team was able to capture nanoscale events that would otherwise remain invisible. This study exemplifies the power of collaborative science. By combining molecular biology, cell imaging, and nanoscale biophysics. For technical details on the PPP-FMR probes used in this study, visit: https://www.nanosensors.com/pointprobe-plus-force-modulation-mode-reflex-coating-afm-tip-PPP-FMR