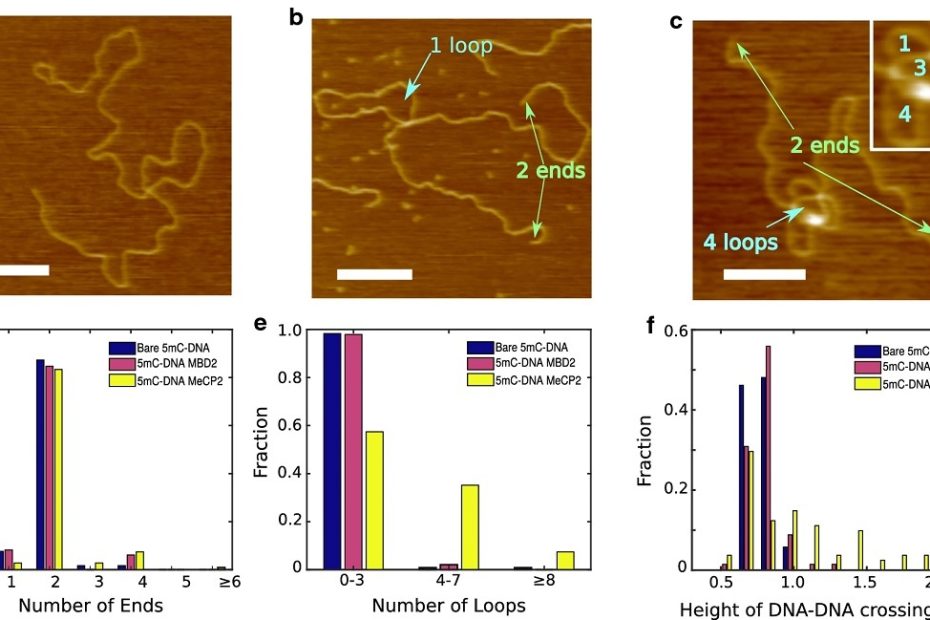
DNA looping by two 5-methylcytosine-binding proteins quantified using nanofluidic devices
MeCP2 and MBD2 are members of a family of proteins that possess a domain that selectively binds 5-methylcytosine in a CpG context. Members of the… Read More »DNA looping by two 5-methylcytosine-binding proteins quantified using nanofluidic devices